Dr. Philip Yangyuoru
Associate Professor
contact information:
Office Hours: :
MW 10 am and 11 am and F 3 pm. All other office hours by appointment.

Research Interest
- Bioanalytical chemistry, biosensors, chemical separations, and forensic biochemistry.
- Forensics and trace analysis of heavy metals in food, water, biological and environmental samples.
- Forensic toxicology and analysis of medicinal plant drug candidates.
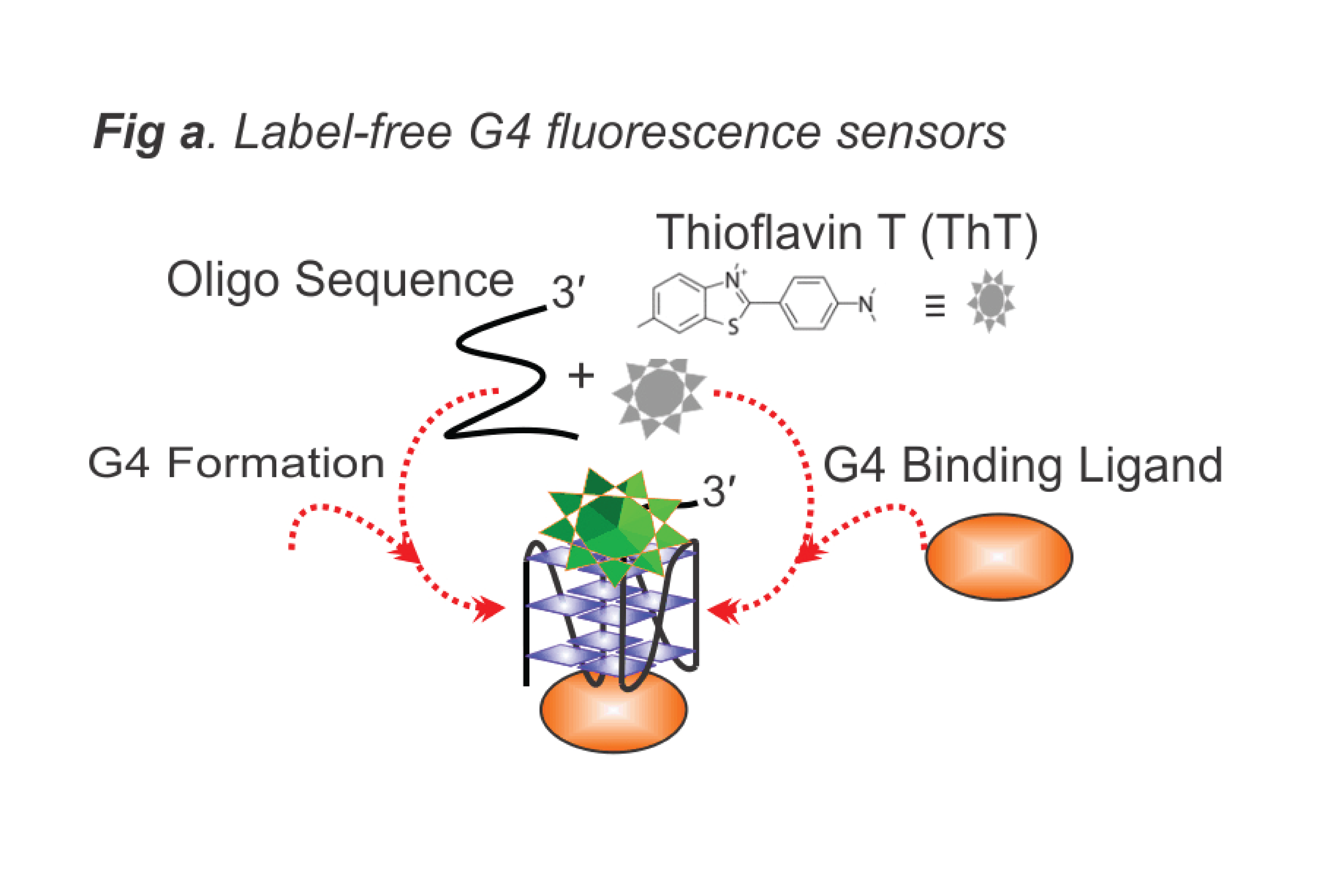
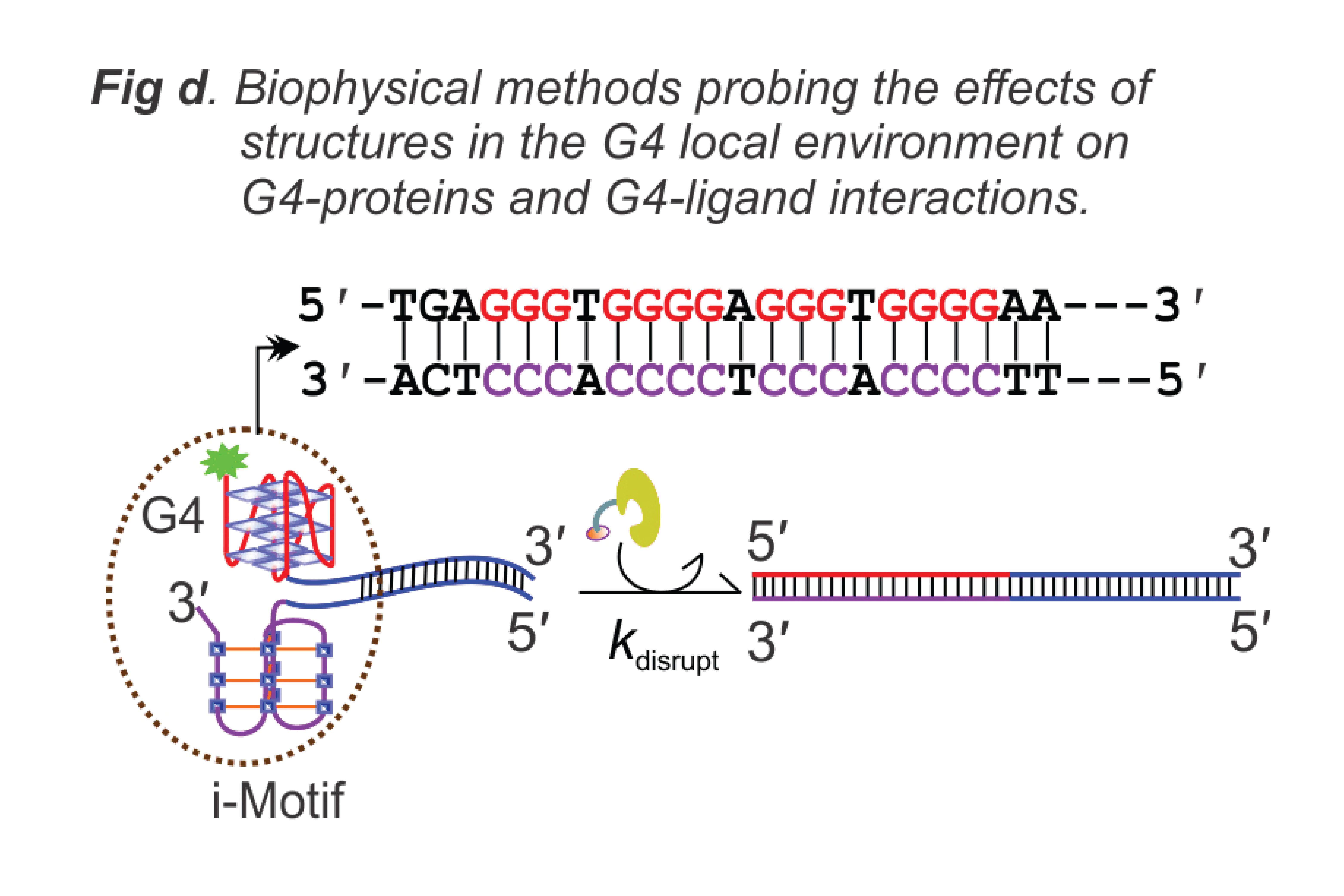
- Develop high-throughput methods for probing noncanonical nucleic acid secondary structures especially G-quadruplexes (Fig. a) and i-motifs (Fig. d).
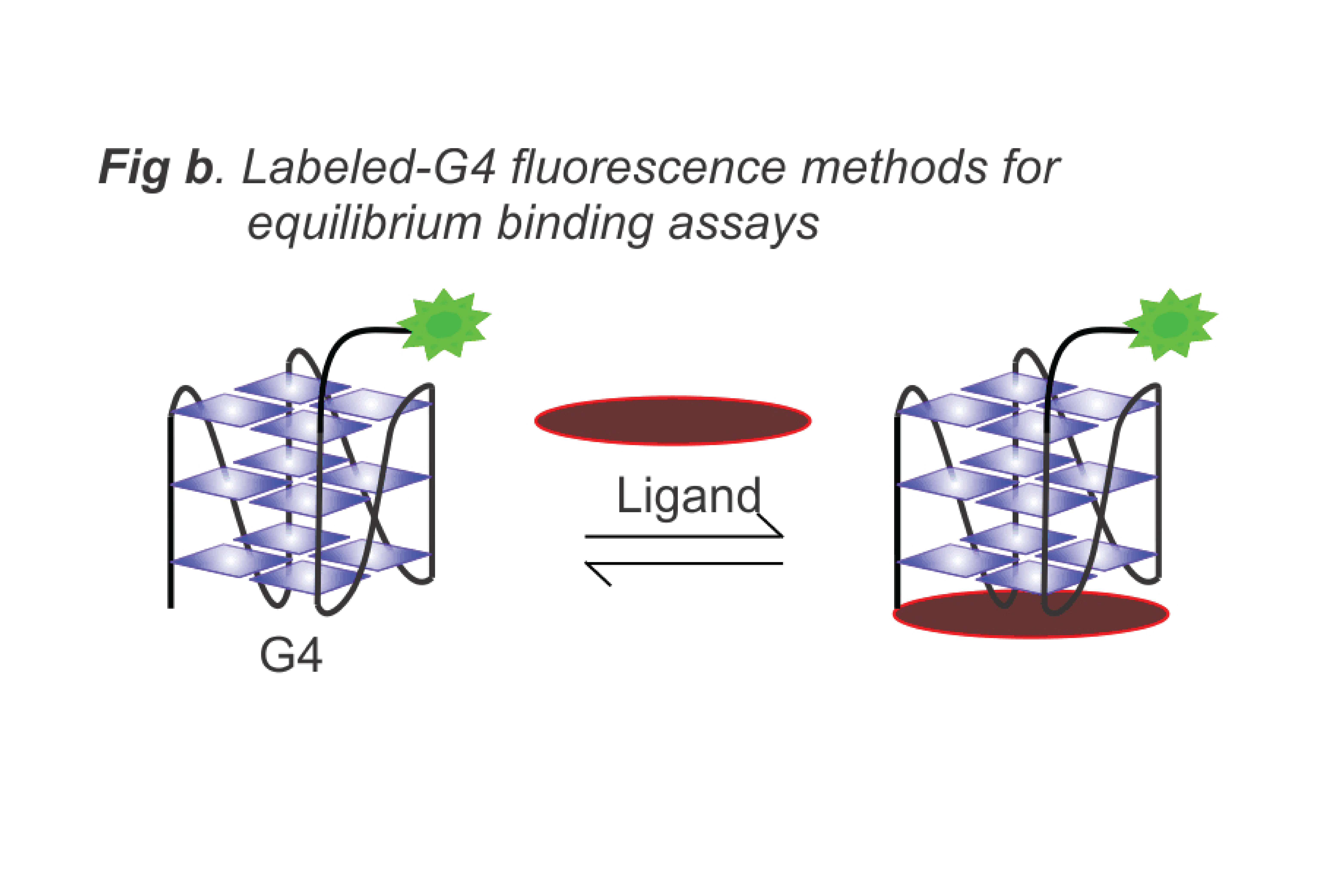
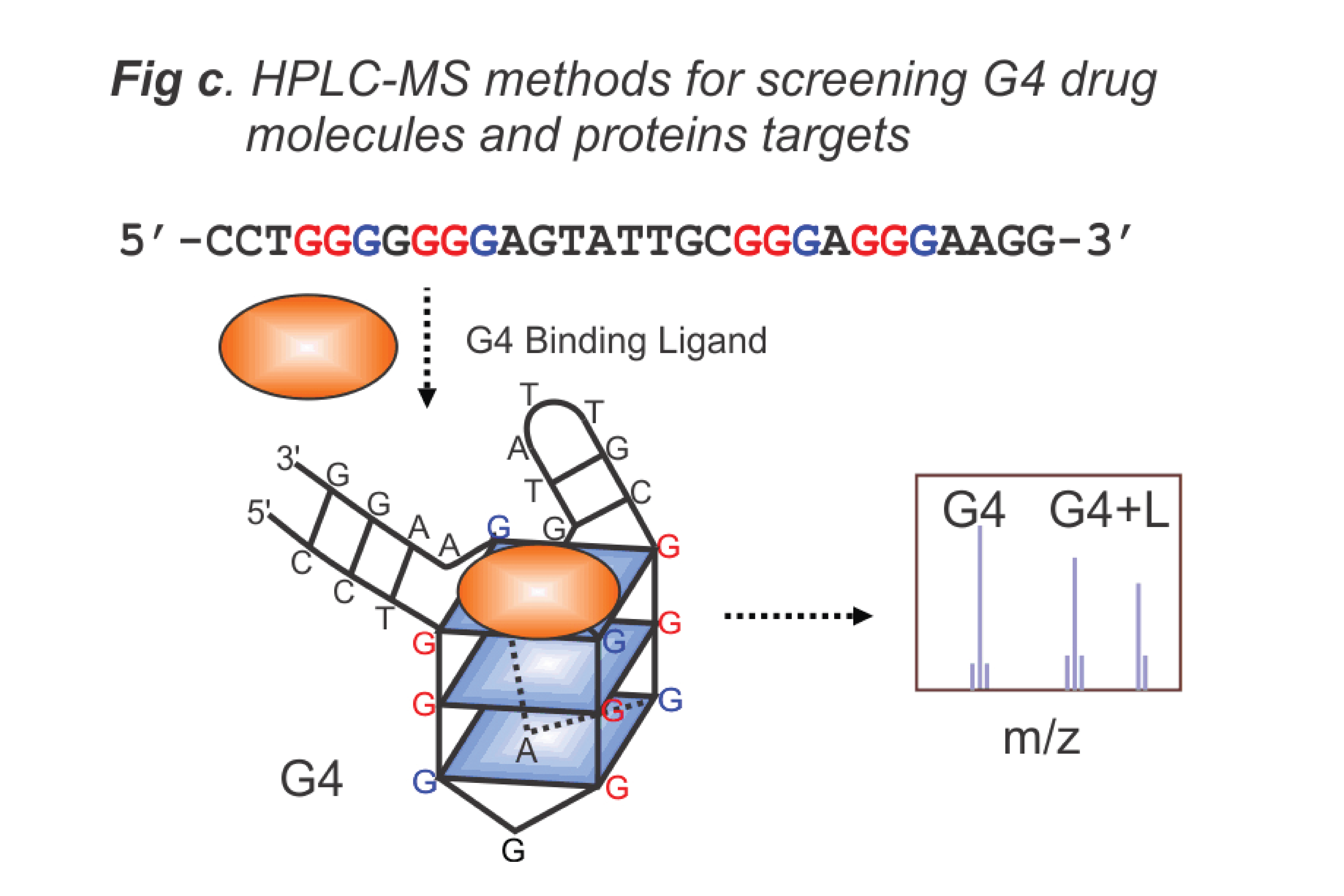
- Design nucleic acid-based biosensors for drug molecules (Fig. b), including HPLC-MS methods for proteins and ligand binding (Fig. c), and for detecting biomarkers of biological and environmental concern.
- Enzyme kinetics assays and mechanisms by which helicase proteins manipulate nucleic acid structures (Fig. d).
Nucleic acid sequences fold into diverse unique secondary structure conformations under different conditions. Single-stranded DNA (ssDNA) regions consisting of at least four closely spaced runs of three or more consecutive guanines or cytosines strongly tend to fold into stable G-quadruplex (G4) or i-motif structures respectively. The stability of these structures makes them suitable for designing biosensors. Moreover, these unique structures have been implicated in regulating many important biological processes.
The analytical instrumentations/methods used for research include fluorescence, electrophoresis, UV/Vis, HPLC/MS, GC/MS, and ICP/MS.
Publications:
1. Edwards AN, Iannucci AN, VanDenBerg J, Kesti A, Rice T, Sethi S, Dhakal S, Yangyuoru PM, "The G-quadruplex structure in the ATP-binding DNA aptamer strongly modulates ligand binding activity". ACS Omega, 2024; doi.org/10.1021/acsomega.3c10386.
2. Chang-Gu B, Bradburn D, Yangyuoru PM, Russell R, “The DHX36-specific-motif (DSM) enhances specificity by
accelerating recruitment of DNA G-quadruplex structures” Journal of Biol. Chem. 2020; doi.10.1515/hsz-2020-0302.
3. Yangyuoru PM, Bradburn D, Liu Z, Xiao TS, Russell R “The G-quadruplex (G4) resolvase DHX36 efficiently and
specifically disrupts DNA G4s via a translocation-based helicase mechanism.” Journal of Biol. Chem. 2017; DOI:
10.1074/jbc.M117.815076.
4. Yangyuoru PM, Di Antonio M, Ghimire C, Biffi G, Balasubramanian S, Mao H, “Dual Binding of an Antibody and a
Small Molecule Increases the Stability of TERRA G-Quadruplex” Angew. Chem. Int. Ed. 2014; DOI:
10.1002/anie.201408113.
5. Yangyuoru PM, Zhang AYQ, Zhe S, Koirala D, Balasubramanian, S Mao H, “Mechanochemical Properties of Individual Human Telomeric RNA G-quadruplexes” ChemBioChem. 2013; DOI: 10.1002/cbic.201300350.
6. Yangyuoru PM, Dhakal S, Yu Z, Koirala D, Mwongela SM, Mao H. “Single-Molecule Measurements of the Binding
between Small Molecules and DNA Aptamers.” Anal Chem. 2012; doi: 10.1021/ac300427d.
7. Yangyuoru PM, Otieno AC, Mwongela SM. “Determination of Sphingosine Kinase 2 Activity using Fluorescent
Sphingosine by Capillary Electrophoresis.” Electrophoresis. 2011; doi: 10.1002/elps.201000495.
8. Yangyuoru PM, Webb JW, Shaw CF III. “Proton-Linked bi- and tri-Metallic Gold Cyanide Complexes observed by ESI-MS Spectrometry.” J. Inorg. Biochem.2008; doi: 10.1016/j.jinorgbio.2007.10.020.
9. Yangyuoru PM, Webb JW, Shaw CF III. “Glutathionato-S-Gold(III) Complexes formed as Intermediates in the
Reduction of Auricyanide by Glutathione.” J. Inorg. Biochem. 2008; doi: 10.1016/j.jinorgbio.2007.10.028.
10. Cui Y, Koirala D, Kang H-J, Dhakal S, Yangyuoru PM, Hurley LH, Mao H. “Molecular Population Dynamics of DNA
Structures in a Bcl-2 Promoter Sequence is Regulated by Small Molecules and the Transcription factor hnRNP LL”
Nucleic Acids Res. 2014; doi: 10.1093/nar/gku185.
11. Punnoose JA, Cui Y, Koirala D, Yangyuoru PM, Ghimire C, Shrestha P, Mao H. “Interaction of G-quadruplexes in the
Full-length 3' Human Telomeric Overhang”, JACS, 2014, 18062-18069. DOI 10.1021/ja510079u.
12. Ghimire C, Park S, Iida K, Yangyuoru PM, Otomo H, Yu Z, Nagasawa K, Sugiyama H, and Mao H, “Direct Quantification of Loop Interaction and π-π Stacking for G-quadruplex Stability at the Submolecular Level”, JACS, 2014,
dx.doi.org/10.1021/ja503585h.
13. Dhakal S, Cui Y, Koirala D, Ghimire C, Kushwaha S, Yu Z, Yangyuoru PM, Mao H. “Structural and Mechanical
Properties of Individual Human Telomeric G-quadruplexes in Molecularly Crowded Solutions” Nucleic Acids Res. 2013;
doi: 10.1093/nar/gkt038.
14. Koirala D, Yangyuoru PM, Mao H. “Mechanical Affinity as a New Metrics to Evaluate Binding Events” Rev. Anal. Chem. 2013: Invited Review. DOI 10.1515/revac-2013-0004.
